Imaging of Plasmids in Liquid using True Non-Contact Mode Atomic Force Microscopy
- 30 May 2017
- Volume 10
- NanoScientific Magazine, Summer 2017
John Paul Pineda, Gerald Pascual, Cathy Lee, Byong Kim, and Keibock Lee
Park Systems Inc., Santa Clara, CA, USA
INTRODUCTION
Deoxyribonucleic acid (DNA) is one of the most significant discoveries of the last century, primarily because it revolutionized many fields including genetics, molecular biology, medical research, agriculture, forensic science, and many others. DNA is a biological molecule that acts as the storage of genetic information essential for bodily function, growth, and the reproduction of all living organisms. This information is located in the nucleus of every cell in an organism and can be transmitted not only from one cell to another but also from parents to offspring. Hence, knowledge on the structure of DNA is very valuable in understanding how the transfer of genetic information occurs.
Several techniques have been used in studying the structure of DNA, one of which is electron microscopy (EM). However, several issues on sample preservation have arisen with this technique due to its tedious sample preparation requirements. As a result, the atomic force microscope (AFM) was developed to overcome such shortcomings. AFM’s capability to image samples under in-liquid conditions preserves the condition of biological samples and provides higher quality high-resolution images [1]. The True Non-Contact imaging mode of AFM tools from Park Systems allows for the measurement of sample topography without requiring a tip-sample interaction that can lead to sample damage and tip degradation. Thus, this mode is preferred when measuring samples that are sensitive to surface deformation rather than conventional contact and tapping modes. The advanced Z scanner design of a Park AFM is the key feature in achieving the non-contact imaging performance that defines Park’s True Non-Contact mode: the highest imaging resolution and most accurate topographical data whilst maintaining tip sharpness and minimizing the chance of damaging the sample surface [2].
EXPERIMENTAL
A 45 ng sample of plasmids (a type of small DNA molecule) was dispersed on a mica substrate and imaged with a Park NX10 AFM under in-liquid conditions using True Non-Contact mode. A cantilever with a relatively low nominal resonance frequency of 110 kHz and a nominal spring constant of 0.09 N/m was utilized in this experiment.
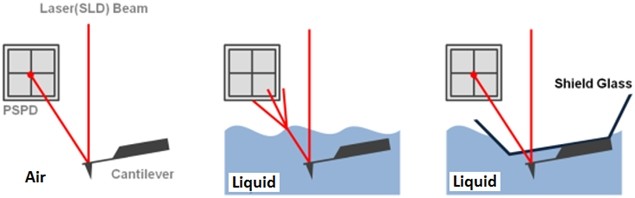
The non-contact AFM image is acquired by measuring the changes in the vibrational amplitude of the cantilever induced by the attractive van der Waals force as the cantilever is mechanically oscillated near its resonant frequency during the scan. The measured changes are compensated for by the AFM’s feedback loop which maintains the cantilever’s constant amplitude and distance. Non-contact mode measures the topography of the sample surface by using this feedback mechanism to control the Z scanner movement [2]. In liquid conditions, the cantilever’s resonant frequency decreases by 1/3 of its original value in air due to dampening effects; a similar decrease is observed in its amplitude as well. In addition, multiple peaks can also be observed on a frequency sweep in liquid—fewer fluctuations are observed in air. Thus, the appropriate resonant frequency was chosen before the measurement procedure by selecting the highest peak near the region around 1/3 of the original resonant frequency and the amplitude was set to around 0.9 nm. The feedback system of the AFM uses a superluminescent diode (SLD) beam reflected from the cantilever. However, due to the instability of the liquid surface during measurement, the reflected SLD beam will scatter. For this reason, a liquid probehand with shielded glass was designed by Park Systems to avoid this problem. This type of probehand was utilized in the experiment.
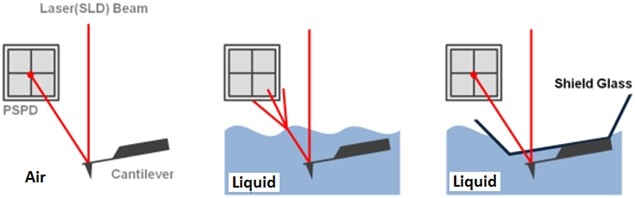
Figure 1. The feedback mechanism of True Non-Contact mode AFM in air and in liquid scan conditions. The shielded probehand in the rightmost image demonstrates its utility in maintaining optimal SLD beam and PSPD performance.
RESULTS & DISCUSSION
A high resolution image of the plasmid sample was successfully acquired and processed using XEI software developed by Park Systems. The sample was expected to consist of uniform linear plasmids. However, the topography data in Figure 2 revealed that the 1 by 1 um scanned region contains a mixture of linear and supercoiled plasmid DNA strands. The strands with larger structures are marked with green arrows and exhibit a supercoiled appearance with diameters of approximately 19 nm while the strands with smaller, linear structures are marked with yellow arrows and appeared to be in a relaxed state with diameters of approximately 8 nm. It was also observed that the supercoiled DNA strands appeared to be longer when compared to the linear strands. The line profile that generated by XEI in Figure 3 provides the height information of DNA strands. The data shows the supercoiled strands have a height of 1.6 nm whereas the height of the linear strand was only 1 nm.
In its normal environment (cell nucleus), the DNA twist itself to overcome distortion as it is subjected to torsional stress during replication. A similar phenomenon could have happened in the linear DNA during this experiment. It is speculated that the linear DNA became supercoiled due to the torsional stress during sample preparation. The end of the plasmid possibly stuck to the substrate more tightly than expected, and as the DNA is subjected to torsional tension, the DNA twists around and bunches up on itself resulting to a larger and supercoiled structure with increasing orders of twisting [3].
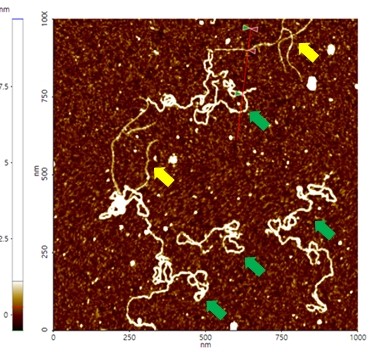
Figure 2. Non-contact mode AFM topography image of plasmids. The segments indicated with yellow arrows have linear structures whereas those with green arrows exhibit supercoiled structures. Scan size: 1 x 1 um.
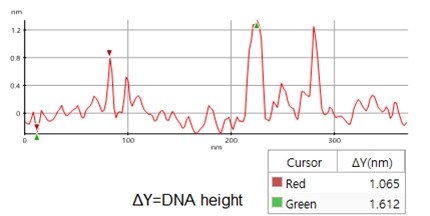
Figure 3. Line profile of non-contact mode AFM topography data taken from the sample shown in Figure 2. Red marker: smaller, linear DNA structure, green marker: larger, supercoiled DNA structure)
SUMMARY
The structure of a plasmid sample was successfully characterized by the Park NX10 AFM using True Non-Contact mode imaging under in-liquid conditions. The topography data revealed that the sample is characterized by the presence of DNA with two different superstructures: a linear configuration and a supercoiled one speculated to be due to torsional tension during sample preparation. One linear plasmid was revealed to have a width of 8 nm and a height of 1 nm. By comparison, one of the supercoiled plasmids had a width of 19 nm and a height of 1.6 nm. Overall, the technique described in this study will successfully provide researchers nanoscale information that is significant in monitoring the effects of structural damage and conformational change in DNA.
REFERENCES
[1] A. Baro, et al., Atomic Force Microscopy in Liquid: Biological Applications. Wiley-VCH, Pages 233-237.
[2] J. Pineda, et al., True Non-Contact Imaging of Various Samples, Retrieved January 12, 2017, from http://www.parkafm.com/index.php/medias/nano-academy/articles
[3] H. Hansma, et al., Reproducible Imaging and Dissection of Plasmid DNA Under Liquid with the Atomic Force Microscope, AAAS, Pages 1180-1181.