Using Atomic Force Microscopy in Medical Research
- 01 Feb 2020
- Volume 19
- NanoScientific Magazine, Spring 2020

Ana Maria Zaske PhD
Internal Medicine, Cardiology Division. University of Texas Health Science Center at Houston. 1881 East Rd, Houston TX, 77054, USA.
Email: Ana.M.Zaske@uth.tmc.edu
Biography
Ana-Maria Zaske graduated in 2001 with a PhD from the University of Manchester Institute of Science and Technology in the UK, where she specialized in high resolution Atomic Force Microscopy (AFM) to resolve bio-enzymatic processes. She used AFM as a Research Associate in the laboratory of Molecular and Cellular Neurobiology, at the UNAM Mexico, to characterize membrane receptors in follicular cells. In 2004 she did her post-doctoral studies at North Carolina State University. In 2008, she joined the University of Texas Health Science Center at Houston, and later she became the manager of the IM Bioscope 2 - UT core facility. Over the course of her career, Dr. Zaske has gained relevant expertise in the principles and applications of AFM in the biological and medical field. She visualizes the AFM technique as a revolutionary tool to explore the nano-world.
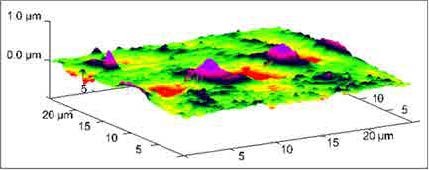
Figure 1. Liposome endocytosis occurring at 60 min of incubation in HCAEC’s. 3D AFM micrograph obtained at 25 μm (x-y) illustrating the engulfment of liposomes conjugated with gold nanoparticles during membrane internalization [1].
Nano-biotechnologies represent an unprecedented recent advance that may revolutionize many areas of medicine and biology. We are now able to evaluate the effects of a treatment in cancer cells, analyze the efficiency of skin healing or just simply observe DNA structures, all at the nano-metric scale. One of the major advances, in the nano-technology field, was the development of the Atomic Force Microscope (AFM) in 1982, as a modification of the first scanning tunneling microscope created by Gerd Binnig. Binnig is the German physicist that won the Nobel Prize in Physics along with Heinrich Rohrer in 1986 for their invention.
The AFM is a nano-technique based on sensing. Such as "feeling" or "touching" the sample surface with a very small flexible probe, while a feedback loop regulates the gap distance between the sample and the probe. The AFM imaging is, therefore, a mechanical process based on attractive and repulsive forces between the sample surface and the probe, giving resolutions to the nanometer level.
Several models of Atomic Force Microscopes have been specifically designed to address the needs of biological and medical investigations. The mission at our AFM core facility located in the University of Texas Health Science Center at Houston, is to explore all meaningful ways to assess experimental trials to mitigate diseases using Atomic Force Microscopy. The BioScope™ II microscope at our core, has been engineered to facilitate advanced life science research such as imaging, probing and manipulating biological systems. All three axes (X, Y, and Z) of the instrument are equipped with high-resolution capacitive sensors to allow imaging at the nanoscale and operating in liquids. It has the ability to analyze samples in the natural state in dry conditions or ‘in-vitro’, immersed in biological solutions. The instrument also has an unprecedented compatibility with optical microscopy to facilitate the localization of the areas of interest.
Cell cultures are widely used to mimic biological conditions where physiological reactions take place. The effects of a treatment or disease can be assessed analyzing the changes in structure and elasticity of living cells using AFM. The interaction of biomolecules with various types of controlled nano-particles is also becoming highly important for many biological and biomedical applications. In combination with fluorescence microscopy, AFM can identify biomolecules based in fluorescence labeling and topography imaging. In addition, non-cellular structures can also be imaged. We can now determine the micromechanical properties of both biological and non-biological materials. Additionally, this technology requires minimal sample preparation procedures, which enhances its popularity in the field.
Some of the AFM applications that we routinely use in our laboratory include:
• Topographical imaging of samples in air or liquid environments
• High-resolution imaging at the nano-metric scale
• Time-lapse experiments that show real-time changes in sample morphology or structure
• Nano-probing of samples to measure molecular interactive forces
• Studies of local micromechanical properties (elasticity, stiffness,
adhesion, roughness)
• Data analysis for determination of homogeneity of samples, size distribution, position, force volume mapping and 3D imaging.
Surface processes such as endocytosis/ exocytosis, interactions between cells and other surfaces, the dynamic reorganization of the cytoskeleton, or analysis of surface configurations are between the topics extensively studied using the AFM technique. An area of major interest, among our researchers, is to determine the stiffness of a sample (Elastic Modulus). By calculating the Young’s modulus on the surface, we can evaluate the effects of several physiological processes by measuring the elastic response on cells and tissues. As per example, the elasticity of the cell membrane can vary between cell types as a function of growth, differentiation, disease or treatment.
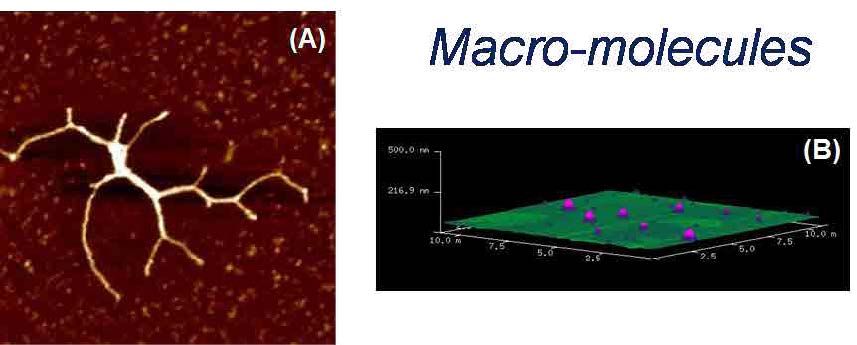
Figure 2. (A) AFM image of a full length myosin protein visualized at RT° on mica. Scan to 2 μm (x-y) using tapping mode in air. (B) 3D AFM micrograph of DPPC liposomes deposited on mica.
Over the years, we have gained relevant experience to apply the fundamentals of AFM to medical research. We initiated studying the internalization mechanism and effects of nanovectors in ‘in-vitro’ cells [1]. We determined the time it takes for the cell to uptake gold labelled liposomes for medical applications. We excitedly captured the moment during liposome membrane internalization as seen in Figure 1. We also studied the cell intake of a multidelivery system consisting of porous silicon microparticles (S1MPs) loaded with one or more types of second-stage nanoparticles. Each level of complexity presents an opportunity for overcoming physiological barriers, such as enzymatic degradation, vascular transport, crossing endothelium, and bypassing molecular efflux pumps [2]. We have also used AFM to determine the structural properties of several other nanovectors such as exosomes, liposomes, microvesicles and chitosan nanoparticles with therapeutic properties (Figure 2).
I have had the opportunity to participate in several multidisciplinary projects at UT-Health. For example, we studied the effects of frozen plasma as a resuscitative agent with Kozar’s group [3]. We determined that plasma based resuscitation, preserved endothelial integrity in an in-vitro model of shock. In these studies, shock (or endothelial injury) disrupted junctional integrity and increased permeability in cell cultures. Fresh frozen plasma restored the endothelial integrity and reduced syndecan-1 shedding after hemorrhagic shock. In a different project, we monitored dynamic morphological changes of human platelets in realtime using AFM. We noticed that platelet formation is independent of the interaction between fibrinogen and the platelet integrin αIIbβ3. However, platelets were very sensitive to agonistinduced micro-vesiculation [4]. In a most recent work with Dr. Yong Zhou [5], AFM was used to determine roughness of cell plasma membranes to quantify several modulations of the lipid-anchored K-Ras expression, which is known to regulate proliferation in cancer cells.
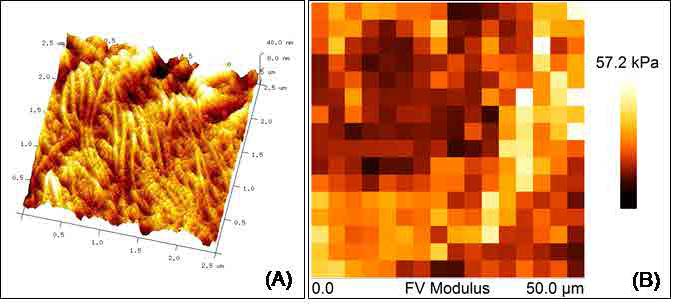
Figure 3. Elastic properties associated with skin healing. Color-coded force maps were generated over skin biopsies to determine collagen deposition as a promoter of wound healing in transgenic mice. (A) AFM micrograph obtained at 2.5 μm (x-y) showing topography of a skin wound. (B) Force Volume map of a 50 μm (x-y) scan of the same skin area.
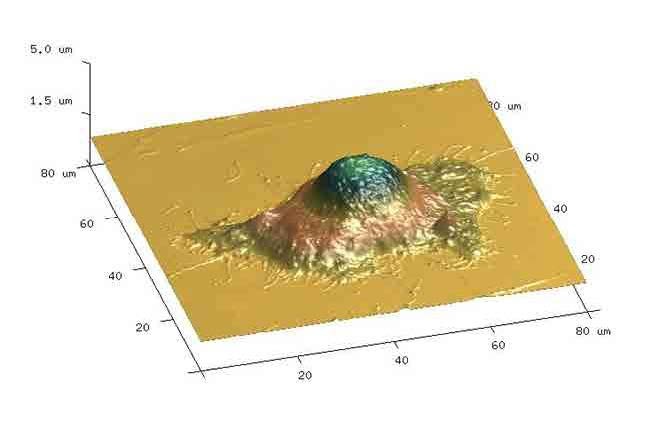
Figure 4. Studies on impact of sucrose as a modulator on cell proliferation and death. Atomic force micrograph of HeLa cells showing surface topography for volume measurements. Cell volume was calculated using the bearing analysis application and using contact mode operated in liquid [8].
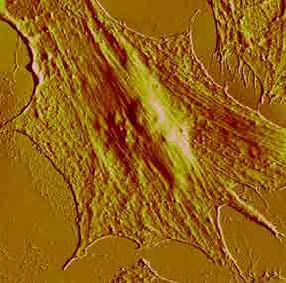
Figure 5. AFM image of a HUVEC (human umbilical vein endothelial cell) expressing E-selectin and incubated with ESTA-1 aptamer nano-beads after fixation with 4% formaldehyde. Image acquired in phosphate saline solution 1X.
New insights have developed, in the AFM field, to determine the elastic properties of biological materials. We routinely estimate the Young’s Modulus of cell membranes, fresh tissues and other biological samples, to help to understand the mechanisms of several physiological processes, such as differentiation, growth, drug treatment, metastasis and disease [6], [7]. Additionally, it is possible to map the distribution of elastic responses on the sample surface (Force Volume maps) by combining force curve measurements with topographic imaging as shown in Figure 3 with tissue samples. The unique properties of the AFM offer unlimited applications in medical research. It can provide valuable insights about the mechanisms of a disease, and assess a successful treatment pathway for its cure.
References
1. ZASKE, ANA-MARIA; DELIA DANILA; MICHAEL C. QUEEN; EVA GOLUNSKI; JODIE L. CONYERS (2013). “Biological Atomic Force Microscopy for Imaging Gold-Labeled Liposomes on Human Coronary Artery Endothelial Cells”. Journal of Pharmaceutics, vol. 2013, Article ID 875906, 8 pages, DOI:10.1155/2013/875906.
2. SERDA, RITA E.; AARON MACK; MERLYN PULIKKATHARA; ANA-MARIA ZASKE; CIRO CHIAPPINI; JEAN R. FAKHOURY; DOUGLAS WEBB; BIANA GODIN; JODIE L. CONYERS; XUE W. LIU; JAMES A. BANKSON; MAURO FERRARI (2010). "Cellular association and assembly of a multi-stage delivery system". Small [cover] 6 (12): 1329-1340.
3. HAYWOOD-WATSON, RICKY; JOHN B. HOLCOMB; ERNEST A. GONZALEZ; ZHANGLONG PENG; SHIBANI PATI; PYONG WOO PARK; WEIWEI WANG; ANA MARIA ZASKE; TYLER MENGE; ROSEMARY A. KOZAR (2011). “Modulation of Syndecan-1 Shedding after Hemorrhagic Shock and Resuscitation”. PLoS ONE 6:8, 1-10, e23530, DOI:10.1371/journal. pone.0023530.
4. ZHANG, YANJUN; XIAO LIU; LI LIU; ANA-MARIA ZASKE; ZHOU ZHOU; YUANYUAN FU; XI YANG; JODIE L. CONYERS; JING-FEI DONG; JIANNING ZHANG (2013). “Contact and Agonist Regulated Microvesiculation of human platelets”. Thrombosis and Haemostasis, 2013; 110 (2):331-339.
5. HONG LIANG, HUANWEN MU, FRANTZ JEANFRANCOIS, BINDU LAKSHMAN, SUPARNA SARKARBANERJEE, YINYIN ZHUANG, YONGPENG ZENG, WEIBO GAO, ANA MARIA ZASKE, DWIGHT NISSLEY, ALEMAYEHU GORFE, WENTING ZHAO, and YONG ZHOU (2019). “Membrane curvature sensing of the lipid-anchored K-Ras small GTPase”. Life Science Alliance (2019) DOI: https://doi.org/10.26508/ lsa.201900343 vol 2 / no 4 / e201900343.
6. LEE, SEI-YOUNG; ANA-MARIA ZASKE; TOMMASO NOVELLINO; DELIA DANILA; MAURO FERRARI; JODIE CONYERS; PAOLO DECUZZI (2011). "Probing the mechanical properties of TNF-α stimulated endothelial cell with atomic force microscopy". International Journal of Nanomedicine 6: 179–195.
7. HONG JIA, JAGADEESH JANJANAM, SHARON C. WU, RUISHAN WANG, GLENDIN PANO, MARINA CELESTINE, OPHELIE MARTINOT, HANNAH BREEZE-JONES, GEORGIA CLAYTON, CECILE GARCIN, ABBAS SHIRINIFARD, ANA MARIA ZASKE, DAVID FINKELSTEIN, MYRIAM LABELLE (2019). “The tumor cell–secreted matricellular protein WISP1 drives pro-metastatic collagen linearization”. The EMBO Journal (2019) E101302. DOI: 10.15252/ EMBJ.2018101302.
8. PULIKKATHARA, MERLYN; COLETTE MARK; NATASHA KUMAR; ANA MARIA ZASKE; RITA E. SERDA (2017). “Sucrose modulation of radiofrequencyinduced heating rates and cell death". Convergent Science Physical Oncology. 3 (2017) 035001. DOI: 10.1088/2057-1739/aa757b.