How Covid-19 Diagnostic Tests Work
- 01 Feb 2020
- Volume 19
- NanoScientific Magazine, Spring 2020
Colin J. Potter and Euan McLeod
Wyant College of Optical Sciences University of Arizona Tucson, AZ 85721
In the last several months, COVID-19 has challenged countries around the world as governments and health systems struggle to contain the spread of the disease and to mitigate its impact. The worldwide response has been remarkable, with nations implementing policies to limit human-to-human spread, distribute and manufacture hospital resources, and ensure that citizens’ basic needs are met. Of all the measures implemented to combat the COVID-19 pandemic, possibly the most impactful is the rapid and early detection of the virus that causes it, SARSCoV-2, which can inform the need for quarantine, treatment, and contact tracing] of infected individuals (Kochańczyk, 2020).
Testing at the desired scale is no easy task, as we have seen in the US and other countries around the world. The scarcity of available test kits, their accuracy and sensitivity, the delay in receiving results, and the relative complexity of the tests themselves all stand as barriers to widespread testing. The development of new, more sensitive, and specific diagnostic technologies has enabled better testing now than was available for the SARS outbreak in the early 2000s, but there is still significant room for improvement.
RT-PCR as a powerful diagnostic tool
Currently, testing for the SARS-CoV-2 virus in the US and around the world is dependent on the use of an approach called reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) or real-time reverse transcriptase PCR to detect the presence of the virus (CDC, 2020). RT-qPCR is performed on a sample taken from a patient, most often a swab of the patient’s upper respiratory tract. The sample is processed to isolate any genetic material in the form of nucleic acids, including human DNA from the patient themselves and any positive-sense RNA present, the genetic material of the SARSCoV- 2 virus.
In RT-qPCR, this RNA is converted into complementary DNA, and then amplified and detected by fluorescence from a DNA binding dye or by a fluorescent DNA probe that binds to a specific sequence of target DNA. The amplification process requires heating and cooling of the sample in a small chamber, with each cycle doubling the amount of target DNA and thus the amount of florescence. Practically, RT-qPCR has demonstrated a limit of detection of 1 copy of RNA per μL in a 30 μL sample (CDC Diagnostic Panel, 2020). With high specificity (i.e. no false positives) based on the use of known primer strands for SARS-CoV-2 RNA, RTqPCR is a powerful tool for diagnosing viral infections. However, as COVID-19 continues to spread, some aspects of RTqPCR have stood as barriers to its rapid and effective implementation.
Diagnosis of COVID-19 using RT-qPCR typically requires sophisticated laboratory equipment and expertise, resulting in a long delay between testing and obtaining results. During this time, a patient may have already spread the virus to several other people, presenting a major problem in the control of an epidemic. Devices such as ID NOW™ and POCKIT™ are relatively new technologies that utilize insulated isothermal polymerase chain reaction, or iiPCR, to qualitatively detect the presence of pathogen RNA (Tsai, 2014). In these devices, the sample is heated from the bottom, creating a convection current where it cools as it moves up in the chamber, effectively providing the heating and cooling cycles of traditional PCR. While this has enabled rapid, point-of-care diagnostic PCR testing to become a reality, it can only give a qualitative result (the presence or absence of pathogen) and is 10-100 fold less sensitive than conventional RT-qPCR (Zhang, 2016).
Additionally, other indications of infection and immunity such as antibodies that the body produces to defend against the virus cannot be detected by either RT-qPCR or iiPCR, so the virus itself must be sampled to detect infection. Besides antibodies, the body also responds to infection by producing cytokine molecules that trigger an inflammatory response. In severe cases, too many cytokines can result in a potentially lethal cytokine storm. In COVID-19, the viral load in upper respiratory secretions has been shown to vary significantly between patients
(Zou, 2020), indicating that detecting viral RNA alone may not be as effective at diagnosing COVID-19 as a test that can simultaneously detect antibodies and other biomarkers.
The perfect COVID-19 diagnostic test
An ideal test for COVID-19 would accomplish several things. One, it should be able to rapidly detect the presence of the viral particle. Two, it should give us information about any antibodies and cytokines that the patient has produced against the virus. Three, it should be sensitive enough to detect even small quantities of these targets in asymptomatic or early infections. Four, it should be administered at the point of care, preferably in an easy-to-use handheld or wearable device that gives results on the spot. And finally, it should be inexpensive enough to be mass produced for the deployment of millions of tests. Such a device would limit user error and testing variability and ultimately enable rapid identification and containment of new outbreaks.
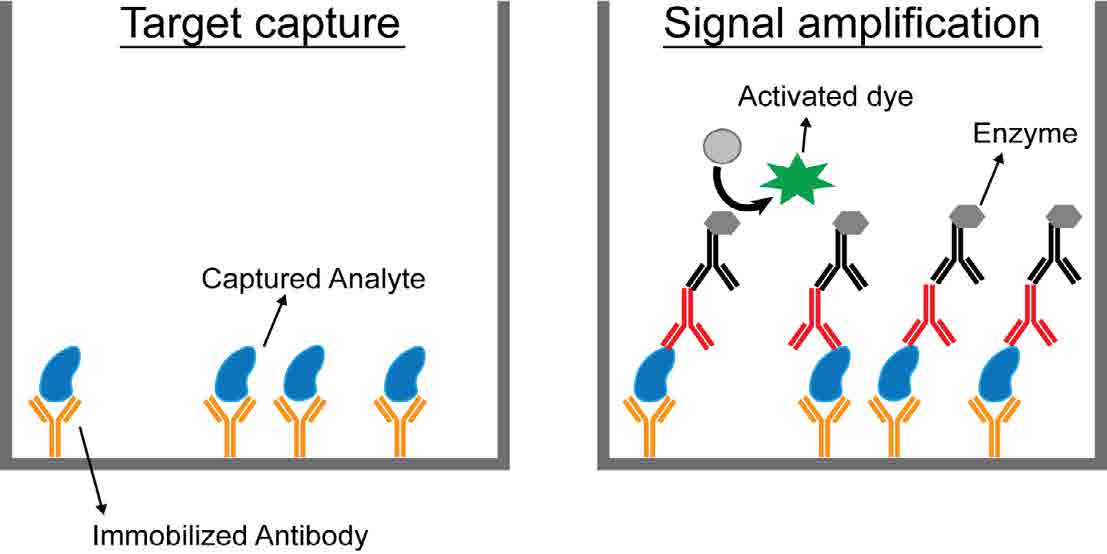
Figure 1: Biosensing using a sandwich ELISA. The target analyte is captured using immobilized antibodies that are reactive against the analyte. All unwanted protein is then washed off before the application of primary and secondary antibodies, which are tagged with enzymes. Signal amplification occurs via enzyme conversion of a dye to a visible form that can be detected and quantified.
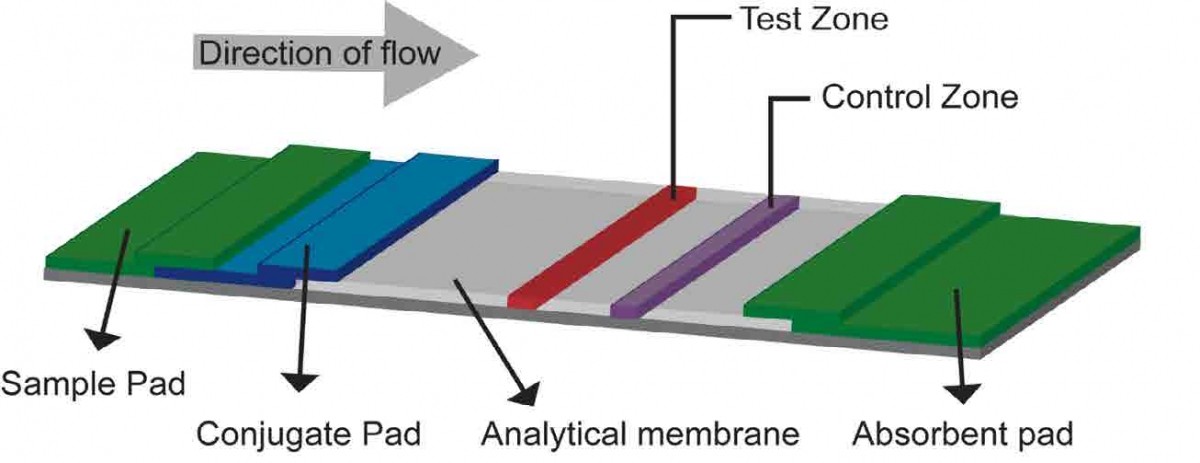
Figure 2: Lateral flow assay (LFA) biosensor. A liquid sample is placed on a sample pad, which then migrates via capillary action to a conjugate release pad, where it can pick up labeled or dye-tagged antibodies that bind specifically to the target analyte to form an antibody-analyte complex. This complex then migrates across a porous membrane towards the detection zone, where a test line containing immobilized anti-analyte antibodies captures the complex, if present. The control zone tests for adequate flow by using another set of antibodies to capture any of the tagged antibodies from the conjugate pad that make it past the test zone.
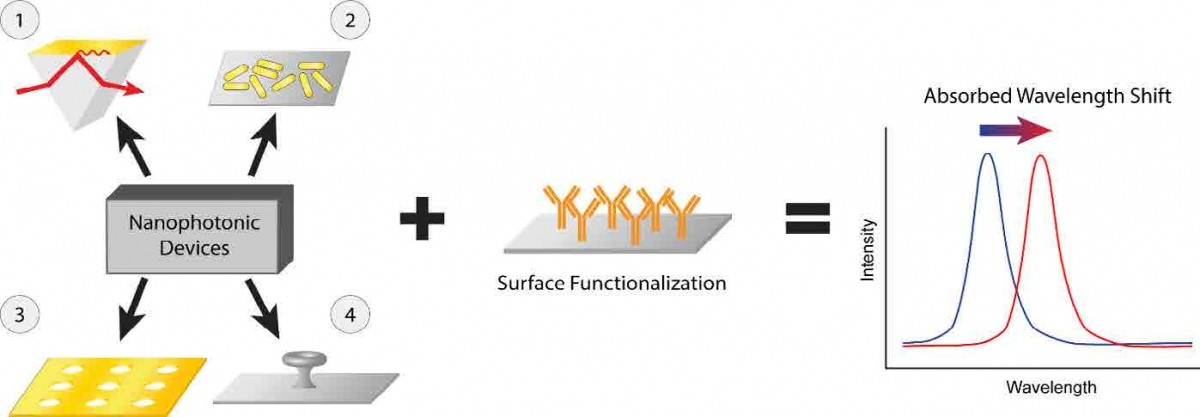
Figure 3: Nanophotonic biosensors. Structures with optical resonances for a particular wavelength of light are fabricated and functionalized with antibodies designed to capture a target molecule. When that target binds to the surface of the sensor, it perturbs the local refractive index, and the resonant wavelength shifts, providing a detectable signal for the sensor. 1- Basic surface plasmon resonance (SPR) detector using gold film on a prism (Liedberg, 1983). 2- Localized SPR, gold nanorod detector (Mayer, 2008). 3- Plasmonic nanohole array (Cetin, 2015). 4- Microtoroid optical resonator (Su, 2016).
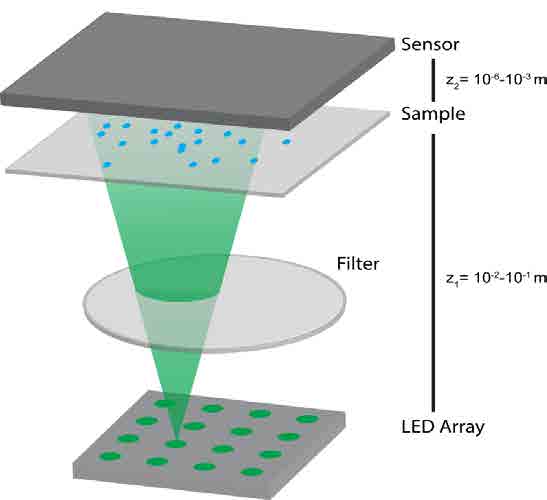
Figure 4: Lens-free holographic microscopy. Light from the LED array illuminates the sample, then is collected after diffracting through objects in the sample field. The sample image is computationally reconstructed to identify particles in the sample plane and generate an image similar to that from a conventional microscope. The ability for this system to be compacted into a handheld device makes it a powerful diagnostic tool.
Detecting beyond RNA with ELISA
One testing method that is already widely used is an enzyme-linked immunosorbent assay (ELISA). In this method, antibodies tagged with enzymes are used to detect target protein molecules in a sample [Fig 1]. This is quite different from PCR, which can only detect DNA or RNA.. Each enzyme can change the color of many dye molecules, resulting in signal amplification, which makes ELISA quite sensitive. While ELISA is not currently widely used to detect SARS-CoV-2 due to the success of RT-qPCR, it still has the potential to be a powerful diagnostic tool in the fight against this and future pandemics.
ELISA has some of the same barriers as PCR when it comes to effective implementation, particularly during an epidemic. ELISA also requires quantitative detection using laboratory machinery with trained staff, and it is more complex than RT-qPCR, requiring controlled storage conditions for the antibodies and leading to similar delays as RT-qPCR from initial testing to results.
Simple point-of-care technologies
Lateral flow assays (LFAs), based on a similar technology as over-the-counter pregnancy tests, can also be used to detect the SARS-CoV-2 virus [Fig 2]. If present, an analyte-antibody complex presents itself as a colored line that indicates a positive test.
The main draw of such a diagnostic technology is that it can be administered and read at the point of care to detect both proteins and RNA/DNA (through use of complementary DNA strands instead of antibodies). Widespread implementation of LFAs could help to control the spread of COVID-19 by providing an instant result. Unfortunately, the main drawback of LFAs is their sensitivity. Due to the design of LFA tests, there is currently no way to amplify a signal, leading to a sensitivity that cannot compete with that of PCR and that is limited by the amount of sample used (Bishop, 2019). Additionally, LFA tests that use new reagents to detect new biomolecules are difficult to produce and optimize, further limiting the usefulness of such technology in the face of a novel pathogen like SARSCoV-2.
Sensing the future
As we continue to push for sensitive, compact, point-of-care diagnostic technologies, the field of nanophotonic biosensors has made great strides in accomplishing these goals. The potential of these sensors makes them attractive as future diagnostic tools. Nanophotonic biosensors utilize unique material structures to confine light to small volumes in order to increase the interaction between the light and the target analyte. Often, nanophotonic biosensors rely on the same DNA or antibody capture molecules used in the previous techniques; however, the detection of the captured molecule is performed optically, which can enable extreme sensitivity in a field-portable package. Devices based on a variety of different material structures have been used that provide limits of detection in the nanomolar to attomolar concentration ranges, including solid metal films (surface plasmon resonance), perforated metal films, arrays of metallic nanoparticles, or glass microstructures where the light skirts their circumference in a whispering gallery mode [Fig 3]. In principle, such technology can be made small and portable, capable of containing all the components necessary for analyte detection in a small chip. These emerging point-of-care technologies are at the forefront of diagnostic biosensing and, with the proper configuration, could be used to detect SARS-CoV-2 surface proteins, RNA, or even antibodies against the virus.
An alternative technology that we are developing employs an emerging form of microscopy called lens-free holographic microscopy [Fig 4]. This technology has the potential to detect a wide dynamic range of analyte concentration, prioritizing both sensitivity and versatility of detectable particles (Xiong, 2018). By using a lensfree system, imaging of this kind can be compacted into a cost-effective hand-held device capable of imaging a sample at sub-micron resolution over a field of view of 20 mm x 20 mm. Reconstruction of the microscopic image is accomplished by computationally processing a recorded diffraction pattern. This enables the detection of nanoscale particles, which can be configured to bind to target biomolecules. This chip-scale sensing approach can form an economically-viable point-of-care diagnostic tool with a high sensitivity to detect the SARS-CoV-2 virus itself as well as the body’s response to the infection.
While there is still some work to be done before these technologies can be implemented on a large scale, we currently stand on the edge of a new era of diagnostic medicine. As new innovations in biomedicine become available, we will be able to detect disease, mitigate the impact of future pandemics, and target personalized medicine using these sensitive and versatile biodetection systems.
Author Biographies:

Colin Potter is an M.D./Ph.D. student studying Optical Sciences in the Wyant College of Optical Sciences at the University of Arizona (UA). He received his B.S. in Neuroscience from UA in 2018 where he studied the effects of parasitic invasion into the brain on the central nervous system. He began studying medicine at the UA College of Medicine Tucson in 2018 and joined the lab of Dr. Euan McLeod to study soft nanophotonic systems and their applications in biodetection.

Euan McLeod is an Assistant Professor in the Wyant College of Optical Sciences at the University of Arizona (UA) since 2015. He is also an Assistant Professor of the UA BIO5 Institute and an Affiliate Member of the UA Cancer Center. He has received postdoctoral training at UCLA and Caltech. He received his Ph.D. from Princeton University and his B.S. from Caltech. Euan’s background and interests lie at the intersection of optics, nanoscience, and soft bio-materials science.
References:
Kochańczyk M, Grabowski F, and Lipniacki T, “Impact of the contact and exclusion rates on the spread of COVID-19 pandemic,” medRxiv (2020).
Centers for Disease Control and Prevention, “CDC Tests for COVID-19,” (2020).
Centers for Disease Control and Prevention, “CDC 2019-Novel Coronavirus (2019 nCoV) Real-Time RT-PCR Diagnostic Panel,” Catalog # 2019-nCoVEUA-01 (2020)
Tsai YL et al., “Validation of a Commercial Insulated Isothermal PCR-based POCKIT Test for Rapid and Easy Detection of White Spot Syndrome Virus Infection in Litopenaeus vannamei,” PLOS One 9, e90545 (2014).
Zhang J et al., “Evaluation of two singleplex reverse transcription- Insulated isothermal PCR tests and a duplex real-time RT-PCR test for the detection of porcine epidemic diarrhea virus and porcine deltacoronavirus,” J Virological Methods 234, 34-42 (2016).
Zou L et al., “SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients,” New England Journal of Medicine 382, 1177–1179 (2020).
Bishop JD, Hsieh HV, Gasperino DJ, and Weigl BH, “Sensitivity enhancement in lateral flow assays: a systems perspective,” Lab on a Chip 19, 2486-2499 (2019).
Soler M, Calvo-Lozano O, Estevez MC, and Lechuga LM, “Nanophotonic Biosensors Driving Personalized Medicine,” Optics and Photonics News, 26-31 (2020).
Leidberg B, Nylander C, and Lundstrom I, “Surface Plasmon Resonance for Gas Detection and Biosensing,” Sensors and Actuators 4, 299-304 (1983).
Mayer KM et al., “A Label-Free Immunoassay Based Upon Localized Surface Plasmon Resonance of Gold Nanorods,” ACS Nano 2, 687-692 (2008).
Cetin AE et al., “Plasmonic Nanohole Arrays on a Robust Hybrid Substrate for Highly Sensitive Label-Free Biosensing,” ACS Photonics 2, 1167-1174 (2015).